Diffraction project datasets PU6423S_3c6c
- Method: SAD
- Resolution: 1.72 Å
- Space group: I 2 2 2
PDB website for 3C6C
PDB validation report for 3C6C
doi:10.18430/M33C6C
Project details
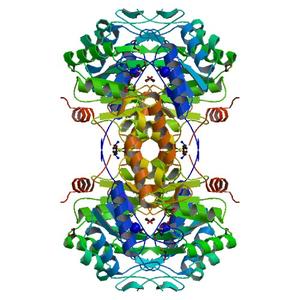
| Title | Crystal structure of a putative 3-keto-5-aminohexanoate cleavage enzyme (reut_c6226) from ralstonia eutropha jmp134 at 1.72 A resolution |
| Authors | Joint Center for Structural Genomics (JCSG) |
| Bioentity | None |
| R / Rfree | 0.15 / 0.18 |
| Unit cell edges [Å] | 50.34 x 121.40 x 133.03 |
| Unit cell angles [°] | 90.0, 90.0, 90.0 |
Dataset 70727_1_###.mccd details

| Number of frames | 180 (1 - 180) |
| Distance [mm] | 250.0 |
| Oscillation width [°] | 0.50 |
| Phi [°] | 177.0 |
| Wavelength [Å] | 0.97964 |
| Experiment Date | 2008-01-12 |
| Equipment | BL9-2 at SSRL (Stanford Synchrotron Radiation Laboratory) |
Dataset 70727_2_###.mccd details
| Number of frames | 90 (1 - 90) |
| Distance [mm] | 400.0 |
| Oscillation width [°] | 1.00 |
| Phi [°] | 177.0 |
| Wavelength [Å] | 0.97964 |
| Experiment Date | 2008-01-12 |
| Equipment | BL9-2 at SSRL (Stanford Synchrotron Radiation Laboratory) |