1469 results
X-ray diffraction data for the Crystal structure of Putative calcium/calmodulin-dependent protein kinase type II association domain (YP_315894.1) from THIOBACILLUS DENITRIFICANS ATCC 25259 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.01 Å
R/Rfree: 0.17/0.21
Resolution: 2.01 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of BH2720 (10175341) from Bacillus halodurans at 1.41 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.41 Å
R/Rfree: 0.17/0.19
Resolution: 1.41 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal structure of the beta subunit of a putative aromatic-ring-hydroxylating dioxygenase (YP_001165631.1) from NOVOSPHINGOBIUM AROMATICIVORANS DSM 12444 at 1.75 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.17/0.21
Resolution: 1.75 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a RNA binding domain of polyadenylate-binding protein (PABPN1) from Homo sapiens at 1.95 A resolution


First author:
Partnership for T-Cell Biology JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.95 Å
R/Rfree: 0.16/0.21
Resolution: 1.95 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the Crystal structure of a branched chain amino acid ABC transporter periplasmic ligand-binding protein (Bxe_C0949) from BURKHOLDERIA XENOVORANS LB400 at 1.88 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.88 Å
R/Rfree: 0.19/0.23
Resolution: 1.88 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the CRYSTAL STRUCTURE OF A HYDROLASE FROM HALOACID DEHALOGENASE-LIKE FAMILY (SP_2064) FROM STREPTOCOCCUS PNEUMONIAE TIGR4 AT 2.10 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.17/0.21
Resolution: 2.10 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a 6-phosphogluconolactonase (Sbal_2240) from Shewanella baltica OS155 at 1.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.40 Å
R/Rfree: 0.17/0.19
Resolution: 1.40 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the CRYSTAL STRUCTURE OF a pyridoxamine 5'-phosphate oxidase-related, FMN binding protein (NPUN_F5749) FROM NOSTOC PUNCTIFORME PCC 73102 AT 1.40 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.40 Å
R/Rfree: 0.17/0.19
Resolution: 1.40 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE GENERAL STRESS PROTEIN 26 (JANN_0955) FROM JANNASCHIA SP. CCS1 AT 2.46 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.46 Å
R/Rfree: 0.23/0.28
Resolution: 2.46 Å
R/Rfree: 0.23/0.28
X-ray diffraction data for the Crystal structure of a Leucine binding protein LivK (TM1135) from Thermotoga maritima MSB8 at 1.90 A resolution


First author:
Joint center for structural genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.15/0.19
Resolution: 1.90 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of a putative nadph-dependent oxidoreductase (dhaf_2064) from desulfitobacterium hafniense dcb-2 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
Resolution: 1.70 Å
X-ray diffraction data for the Crystal structure of a putative cell surface protein (BACCAC_03700) from Bacteroides caccae ATCC 43185 at 2.07 A resolution

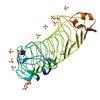
First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.07 Å
R/Rfree: 0.18/0.21
Resolution: 2.07 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of Oligopeptide ABC transporter, periplasmic oligopeptide-binding (TM1223) from THERMOTOGA MARITIMA at 1.73 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.73 Å
R/Rfree: 0.15/0.18
Resolution: 1.73 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the CRYSTAL STRUCTURE OF a pyridoxamine 5'-phosphate oxidase-like family protein (NPUN_R6570) FROM NOSTOC PUNCTIFORME PCC 73102 AT 1.80 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.18/0.20
Resolution: 1.80 Å
R/Rfree: 0.18/0.20
X-ray diffraction data for the Crystal structure of a putative gmp synthase subunit a protein (ta0944m) from thermoplasma acidophilum at 2.45 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.24 Å
R/Rfree: 0.18/0.25
Resolution: 2.24 Å
R/Rfree: 0.18/0.25
X-ray diffraction data for the Crystal structure of a Putative thua-like protein (PARMER_02418) from Parabacteroides merdae ATCC 43184 at 1.75 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.15/0.17
Resolution: 1.75 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the Crystal structure of a diaminohydroxyphosphoribosylaminopyrimidine deaminase/ 5-amino-6-(5-phosphoribosylamino)uracil reductase (tm1828) from thermotoga maritima at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.16/0.20
Resolution: 1.80 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of putative glycosyl hydrolase, was Domain of Unknown Function (DUF1080) (YP_001302580.1) from Parabacteroides distasonis ATCC 8503 at 2.36 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.36 Å
R/Rfree: 0.20/0.25
Resolution: 2.36 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal structure of sugar phosphate isomerase from a cupin superfamily SPO2919 from Silicibacter pomeroyi (YP_168127.1) from SILICIBACTER POMEROYI DSS-3 at 2.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.20/0.25
Resolution: 2.30 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal structure of a DUF4623 family protein (BACEGG_03550) from Bacteroides eggerthii DSM 20697 at 1.91 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.91 Å
R/Rfree: 0.15/0.19
Resolution: 1.91 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of Amino Transferase (RER070207001803) from Eubacterium rectale at 2.10 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.17/0.22
Resolution: 2.10 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of Protein related to general stress protein 26(GS26) of B.subtilis (pyridoxinephosphate oxidase family) (NP_350077.1) from CLOSTRIDIUM ACETOBUTYLICUM at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.19/0.24
Resolution: 2.00 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of a Deoxyribose-phosphate aldolase (TM_1559) from THERMOTOGA MARITIMA at 1.83 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.83 Å
R/Rfree: 0.15/0.19
Resolution: 1.83 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the CRYSTAL STRUCTURE OF A C-TERMINAL FRAGMENT OF THE PUTATIVE FLAGELLAR MOTOR SWITCH PROTEIN FLIN (TM0680) FROM THERMOTOGA MARITIMA AT 1.85 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.17/0.21
Resolution: 1.85 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of the apo form of a 3alpha-hydroxysteroid dehydrogenase (BaiA2) associated with secondary bile acid synthesis from Clostridium scindens VPI12708 at 1.90 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.16/0.19
Resolution: 1.90 Å
R/Rfree: 0.16/0.19