6726 results
X-ray diffraction data for the 1.83 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-404) of Elongation Factor G from Enterococcus faecalis


X-ray diffraction data for the 1.78 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-405) of Elongation Factor G from Haemophilus influenzae


X-ray diffraction data for the 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae.

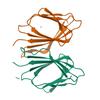
X-ray diffraction data for the Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase in complex with adenine from Vibrio fischeri ES114


X-ray diffraction data for the 1.72 Angstrom Resolution Crystal Structure of 2-Oxoglutarate Dehydrogenase Complex Subunit Dihydrolipoamide Dehydrogenase from Bordetella pertussis in Complex with FAD


X-ray diffraction data for the Crystal structure of peptidase B from Yersinia pestis CO92 at 2.75 A resolution


X-ray diffraction data for the Crystal structure of a fragment (1-405) of an elongation factor G from Vibrio vulnificus CMCP6


X-ray diffraction data for the Crystal structure of the putative periplasmic solute-binding protein from Campylobacter jejuni


X-ray diffraction data for the Structure of spermidine N-acetyltransferase SpeG from Vibrio cholerae


X-ray diffraction data for the 2.0 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Streptococcus pneumoniae in Complex with Uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and Magnesium.


X-ray diffraction data for the Crystal structure of a putative D-alanyl-D-alanine carboxypeptidase from Vibrio cholerae O1 biovar eltor str. N16961


X-ray diffraction data for the 2.05 Angstrom Resolution Crystal Structure of C-terminal Domain (DUF2156) of Putative Lysylphosphatidylglycerol Synthetase from Agrobacterium fabrum.


X-ray diffraction data for the 1.73 Angstrom Resolution Crystal Structure of Dihydropteroate Synthase (folP-SMZ_B27) from Soil Uncultured Bacterium.


First author:
G. Minasov
Gene name: None
Resolution: 1.73 Å
R/Rfree: 0.16/0.19
Gene name: None
Resolution: 1.73 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the X-ray structure of a putative triosephosphate isomerase from Toxoplasma gondii ME49


First author:
E.V. Filippova
Uniprot: A0A125YP67
Gene name: TPI-II
Resolution: 2.00 Å
R/Rfree: 0.17/0.21
Uniprot: A0A125YP67
Gene name: TPI-II
Resolution: 2.00 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Yersinia pestis


X-ray diffraction data for the Structure of the ornithine aminotransferase from Toxoplasma gondii in complex with inactivator


X-ray diffraction data for the Crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 in a complex with (S)-4-amino-5-fluoropentanoic acid


X-ray diffraction data for the Crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 in a complex with gabaculine


X-ray diffraction data for the 1.88 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus mutans UA159 in Complex with FAD


X-ray diffraction data for the 2.15 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis


X-ray diffraction data for the 1.36 Angstrom Resolution Crystal Structure of Malate Synthase G from Pseudomonas aeruginosa in Complex with Glycolic Acid.


X-ray diffraction data for the 1.55 Angstrom Resolution Crystal Structure of Glutathione Reductase from Yersinia pestis in Complex with FAD


X-ray diffraction data for the Crystal structure of the serine endoprotease from Yersinia pestis


First author:
E.V. Filippova
Uniprot: A0A0B6NK33
Gene name: degS
Resolution: 1.85 Å
R/Rfree: 0.15/0.18
Uniprot: A0A0B6NK33
Gene name: degS
Resolution: 1.85 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the 2.65 Angstrom Resolution Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with SAH and NAD


X-ray diffraction data for the 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis.


X-ray diffraction data for the 1.9 Angstrom Resolution Crystal Structure of dTDP-4-dehydrorhamnose Reductase from Yersinia enterocolitica


X-ray diffraction data for the 2.75 Angstrom Resolution Crystal Structure of Acetamidase from Yersinia enterocolitica.


X-ray diffraction data for the 2.45 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Campylobacter jejuni.


X-ray diffraction data for the 1.95 Angstrom Resolution Crystal Structure of Fragment (35-274) of Membrane-bound Lytic Murein Transglycosylase F from Yersinia pestis.


X-ray diffraction data for the 1.95 Angstrom Resolution Crystal Structure of Stage II Sporulation Protein D (SpoIID) from Clostridium difficile in Apo Conformation


X-ray diffraction data for the 1.95 Angstrom Resolution Crystal Structure of Penicillin Binding Protein 2X from Streptococcus thermophilus


X-ray diffraction data for the 1.05 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Acinetobacter baumannii in Covalently Bound Complex with (2R)-2-(phosphonooxy)propanoic Acid.


X-ray diffraction data for the 2.6 Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae


X-ray diffraction data for the 2.3 Angstrom Resolution Crystal Structure of Glutathione Reductase from Vibrio parahaemolyticus in Complex with FAD.


X-ray diffraction data for the 2.22 Angstrom Crystal Structure of N-terminal Fragment (residues 1-419) of Elongation Factor G from Legionella pneumophila.


X-ray diffraction data for the Crystal structure of the Zika virus NS3 helicase.


First author:
S. Nocadello
Uniprot: A0A0X8GJ44
Gene name: None
Resolution: 2.05 Å
R/Rfree: 0.18/0.25
Uniprot: A0A0X8GJ44
Gene name: None
Resolution: 2.05 Å
R/Rfree: 0.18/0.25
X-ray diffraction data for the 1.50 Angstrom Crystal Structure of C-terminal Fragment (residues 322-384) of Iron Uptake System Component EfeO from Yersinia pestis.


X-ray diffraction data for the 2.05 Angstrom Resolution Crystal Structure of Peptidoglycan-Binding Protein from Clostridioides difficile in Complex with Glutamine Hydroxamate.


X-ray diffraction data for the Crystal structure of a GNAT Superfamily PA3944 acetyltransferase in complex with CoA (P1 space group)


X-ray diffraction data for the Succinyl-CoA synthase from Campylobacter jejuni


X-ray diffraction data for the Succinyl-CoA synthase from Francisella tularensis, phosphorylated, in complex with CoA


X-ray diffraction data for the A 2.05A X-Ray Structureof A Bacterial Extracellular Solute-binding Protein, family 5 for Bacillus anthracis str. Ames


X-ray diffraction data for the 1.9 Angstrom Resolution Crystal Structure of Maltose-Binding Periplasmic Protein MalE from Listeria monocytogenes in Complex with Maltose


X-ray diffraction data for the 2.2 Angstrom Resolution Crystal Structure of P-Hydroxybenzoate Hydroxylase from Pseudomonas putida in Complex with FAD.


First author:
L.Shuvalova G.Minasov
Gene name: pobA
Resolution: 2.20 Å
R/Rfree: 0.17/0.22
Gene name: pobA
Resolution: 2.20 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the 1.5 Angstrom Resolution Crystal Structure of NAD-Dependent Epimerase from Klebsiella pneumoniae in Complex with NAD.


X-ray diffraction data for the A 1.85A X-Ray Structure from Peptoclostridium difficile 630 of a Hypothetical Protein


X-ray diffraction data for the 2.35 Angstrom Crystal Structure Minor Lipoprotein from Acinetobacter baumannii.


X-ray diffraction data for the Beta-lactamase penicillinase from Bacillus megaterium


X-ray diffraction data for the Beta-lactamase from Chitinophaga pinensis


X-ray diffraction data for the 2.6 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Aristeromycin and NAD


X-ray diffraction data for the 1.35 Angstrom Resolution Crystal Structure of a Pullulanase-specific Type II Secretion System Integral Cytoplasmic Membrane Protein GspL (N-terminal fragment; residues 1-237) from Klebsiella pneumoniae.


X-ray diffraction data for the 2.15 Angstrom Crystal Structure of N-acetylmuramoyl-L-alanine Amidase from Staphylococcus aureus.


X-ray diffraction data for the 1.95 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenine and NAD


X-ray diffraction data for the 1.8 Angstrom Resolution Crystal Structure of Dimerization and Transpeptidase domains (residues 39-608) of Penicillin-Binding Protein 1 from Staphylococcus aureus.


X-ray diffraction data for the 2.5 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD.


X-ray diffraction data for the Crystal structure of Zika virus NS2B-NS3 protease in apo-form.


X-ray diffraction data for the 1.47 Angstrom Crystal Structure of the C-terminal Substrate Binding Domain of LysR Family Transcriptional Regulator from Klebsiella pneumoniae.


X-ray diffraction data for the 2.8 Angstrom Crystal Structure of the C-terminal Dimerization Domain of Transcriptional Regulator PdhR from Escherichia coli.


X-ray diffraction data for the 2.4 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with DZ2002 and NAD


X-ray diffraction data for the 1.25 Angstrom Crystal Structure of Chitinase from Bacillus anthracis.


X-ray diffraction data for the Crystal Structure of an ABC Transporter Substrate-Binding Protein from Listeria monocytogenes EGD-e


X-ray diffraction data for the 1.0 Angstrom Crystal Structure of pre-Peptidase C-terminal Domain of Collagenase from Bacillus anthracis.


X-ray diffraction data for the Listeria monocytogenes internalin-like protein lmo2027


X-ray diffraction data for the Dehydroquinate dehydratase and shikimate dehydrogenase from S. pombe AroM


X-ray diffraction data for the Listeria monocytogenes OppA bound to peptide

X-ray diffraction data for the X-Ray Crystal Structure of a Fragment (1-75) of a Transcriptional Regulator PdhR from Escherichia coli CFT073


X-ray diffraction data for the Dehydroquinate dehydratase from A. fumigatus AroM


X-ray diffraction data for the Catalytic domain of LPMO Lmo2467 from Listeria monocytogenes


X-ray diffraction data for the 1.9 Angstrom Crystal Structure of NS5 Methyl Transferase from Dengue Virus 1 in Complex with S-Adenosylmethionine and Beta-D-Fructopyranose.


X-ray diffraction data for the Cycloalternan-degrading enzyme from Trueperella pyogenes in complex with covalent intermediate


X-ray diffraction data for the Secreted Internalin-like protein Lmo2445 from Listeria monocytogenes


X-ray diffraction data for the 1.65 Angstrom Crystal Structure of Triosephosphate Isomerase (TIM) from Streptococcus pneumoniae


X-ray diffraction data for the 1.32 Angstrom Crystal Structure of Ybbr like Domain of lmo2119 Protein from Listeria monocytogenes.


X-ray diffraction data for the Cycloalternan-forming enzyme from Listeria monocytogenes in complex with maltopentaose


X-ray diffraction data for the 2.5 Angstrom Crystal Structure of Putative Lipoprotein from Clostridium perfringens


X-ray diffraction data for the 1.35 Angstrom Crystal Structure of C-terminal Domain of Glycosyl Transferase Group 1 Family Protein (LpcC) from Francisella tularensis.


X-ray diffraction data for the 2.6 Angstrom Resolution Crystal Structure of Stage II Sporulation Protein D (SpoIID) from Clostridium difficile in Complex with Triacetylchitotriose


X-ray diffraction data for the Cycloalternan-forming enzyme from Listeria monocytogenes in complex with panose


X-ray diffraction data for the 2.2 Angstrom Crystal Structure of ABC Transporter Substrate Binding Protein CtaP (Lmo0135) from Listeria monocytogenes.


X-ray diffraction data for the 2.85 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenosine and NAD.


X-ray diffraction data for the 2.25 Angstrom Resolution Crystal Structure of Fatty-Acid-CoA Ligase (FadD32) from Mycobacterium smegmatis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine.


X-ray diffraction data for the 1.65 Angstrom resolution crystal structure of lmo0182 (residues 1-245) from Listeria monocytogenes EGD-e


First author:
A.S. Halavaty
Uniprot: Q8YAE8
Gene name: lmo0182
Resolution: 1.65 Å
R/Rfree: 0.16/0.19
Uniprot: Q8YAE8
Gene name: lmo0182
Resolution: 1.65 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the 1.93 Angstrom resolution crystal structure of a pullulanase-specific type II secretion system integral cytoplasmic membrane protein GspL (C-terminal fragment; residues 309-397) from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044


First author:
A.S. Halavaty
Uniprot: A0A060VDE2
Gene name: pulL
Resolution: 1.93 Å
R/Rfree: 0.23/0.28
Uniprot: A0A060VDE2
Gene name: pulL
Resolution: 1.93 Å
R/Rfree: 0.23/0.28
X-ray diffraction data for the Crystal structure of Listeria monocytogenes InlP


X-ray diffraction data for the 2.25 Angstrom Resolution Crystal Structure of Long-chain-fatty-acid-AMP Ligase FadD32 from Mycobacterium tuberculosis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine


X-ray diffraction data for the 1.52 Angstrom Crystal Structure of Fc fragment of Human IgG1.


X-ray diffraction data for the 1.75 Angstrom Crystal Structure of Superantigen-like Protein, Exotoxin from Staphylococcus aureus, in Complex with Sialyl-LewisX.


X-ray diffraction data for the Cycloalternan-degrading enzyme from Trueperella pyogenes in complex with isomaltose


X-ray diffraction data for the Cycloalternan-degrading enzyme from Trueperella pyogenes in complex with cycloalternan


X-ray diffraction data for the A 1.55A X-Ray Structure from Vibrio cholerae O1 biovar El Tor of a Hypothetical Protein


First author:
J.S. Brunzelle
Uniprot: A0A0H7H6M3
Gene name: None
Resolution: 1.55 Å
R/Rfree: 0.17/0.21
Uniprot: A0A0H7H6M3
Gene name: None
Resolution: 1.55 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the 2.6 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) G234S mutant from Staphylococcus aureus (IDP00699) in complex with NAD+ and BME-free Cys289


X-ray diffraction data for the 1.75 Angstrom resolution crystal structure of the apo-form acyl-carrier-protein synthase (AcpS) (acpS; purification tag off) from Staphylococcus aureus subsp. aureus COL in the I4 space group


X-ray diffraction data for the 2.11 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289


X-ray diffraction data for the 2.25 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) Y450L point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289


X-ray diffraction data for the 1.5 Angstrom Crystal Structure of Shikimate Dehydrogenase 1 from Peptoclostridium difficile.


X-ray diffraction data for the 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium


X-ray diffraction data for the Cycloalternan-forming enzyme from Listeria monocytogenes in complex with pentasaccharide substrate


X-ray diffraction data for the 2.1 Angstrom Crystal Structure of Stage II Sporulation Protein D from Bacillus anthracis


X-ray diffraction data for the 2.65 Angstrom Resolution Crystal Structure of an orotate phosphoribosyltransferase from Bacillus anthracis str. 'Ames Ancestor' in complex with 5-phospho-alpha-D-ribosyl diphosphate (PRPP)


X-ray diffraction data for the 2.55 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in complex with shikimate-3-phosphate, phosphate, and potassium

